Today I scouted an issue, which I already have solved at least twice formerly - and couldn't remember at all. Getting older and older...
d.my <- data.frame(cal_d=as.Date(c("2011-12-30","2012-01-03","2012-04-07","2012-07-08")), y=1:4)
The following code would fail to produce a grid():
plot(y ~ cal_d, d.my, panel.first=grid())
This will paint the grid, but choose the wrong ticks:
plot(y ~ cal_d, d.my, panel.first=quote(grid()))
This will paint the grid correctly:
plot(y ~ cal_d, d.my, panel.first=quote({
grid(nx=NA, ny=NULL)
abline(v=axis.Date(1, x=d.my$cal_d, labels=NA, col=NA), col="grey", lty="dotted")
}))
Reference found: https://stat.ethz.ch/pipermail/r-help/2011-May/279169.html
Freitag, 3. Juni 2016
Freitag, 11. März 2016
Methods and Classes
How can we find the defined functions for a special class?
# for which classes does my function provide an own interface?
methods("summary")
# which functions do support my class?
methods(class="matrix")
# get the source code, if not readily available
getAnywhere("Desc.character")
# for which classes does my function provide an own interface?
methods("summary")
# which functions do support my class?
methods(class="matrix")
# get the source code, if not readily available
getAnywhere("Desc.character")
Dienstag, 20. Januar 2015
DescTools approaching v. 1.0...
A next version of DescTools 0.99.16 has been submitted to CRAN today.
It includes a handful new functions and several performance enhancements.
See: http://cran.r-project.org/web/packages/DescTools/NEWS
It includes a handful new functions and several performance enhancements.
See: http://cran.r-project.org/web/packages/DescTools/NEWS
Montag, 24. November 2014
p-values uniformly distributed
Are p-values really uniformly distributed? They are:
library(DescTools)
gg <- rnorm(10000)
n <- 100
m <- sapply(1:n, function(x) sample(1:10000, size = n))
m[] <- gg[m]
d.frm <- data.frame(x=as.vector(m), grp=factor(rep(1:n, each=n)))
p <- na.omit(as.vector(pairwise.t.test(x = d.frm$x, g = d.frm$grp,
p.adjust.method = "none", pool.sd = FALSE)$p.value))
Desc(p)
See: https://www.youtube.com/watch?v=5OL1RqHrZQ8
library(DescTools)
gg <- rnorm(10000)
n <- 100
m <- sapply(1:n, function(x) sample(1:10000, size = n))
m[] <- gg[m]
d.frm <- data.frame(x=as.vector(m), grp=factor(rep(1:n, each=n)))
p <- na.omit(as.vector(pairwise.t.test(x = d.frm$x, g = d.frm$grp,
p.adjust.method = "none", pool.sd = FALSE)$p.value))
Desc(p)
See: https://www.youtube.com/watch?v=5OL1RqHrZQ8
Mittwoch, 2. April 2014
Unique List, but with order
How to create a list of elements in order:
(x <- structure(c(3L, 3L, 3L, 3L, 1L, 2L, 2L,
2L, 2L, 3L, 2L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L),
.Label = c("B", "C", "D"), class = "factor"))
[1] D D D D B C C C C D C B B C C C C C C C C C
Levels: B C D
(x <- structure(c(3L, 3L, 3L, 3L, 1L, 2L, 2L,
2L, 2L, 3L, 2L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L),
.Label = c("B", "C", "D"), class = "factor"))
[1] D D D D B C C C C D C B B C C C C C C C C C
Levels: B C D
> x[c(TRUE, diff(as.numeric(x))!=0)] [1] D B C D C B C Levels: B C D
Montag, 27. Januar 2014
Elegant Summary
A really elegant solution for combining different summary results:
library(plyr)
mtcars$mpg_g <- cut(mtcars$mpg, breaks=seq(from=10, to=35, by=5))
d.res <- ddply(mtcars, c("cyl", "am"), summarize,
anz = length(cyl),
min.hp=min(hp), med.hp=median(hp), mean.hp=round(mean(hp),2), max.hp=max(hp),
mpg_g=matrix(table(mpg_g), nrow=1))
d.res
Credits to Markus Naepflin (2014) for that.
library(plyr)
mtcars$mpg_g <- cut(mtcars$mpg, breaks=seq(from=10, to=35, by=5))
d.res <- ddply(mtcars, c("cyl", "am"), summarize,
anz = length(cyl),
min.hp=min(hp), med.hp=median(hp), mean.hp=round(mean(hp),2), max.hp=max(hp),
mpg_g=matrix(table(mpg_g), nrow=1))
d.res
Credits to Markus Naepflin (2014) for that.
Dienstag, 21. Januar 2014
DescTools 0.99.6 released
The first Version of DescTools can be downloaded from CRAN. So it is available as well in Mac-Version:
http://cran.r-project.org/web/packages/DescTools/index.html
DescTools contains a bunch of basic statistic functions and convenience wrappers for efficiently describing data, creating specific plots, doing reports using MS Word, Excel or PowerPoint. The package's intention is to offer a toolbox, which facilitates the (notoriously time consuming) first descriptive tasks in data analysis, consisting of calculating descriptive statistics, drawing graphical summaries and reporting the results. Many of the included functions can be found scattered in other packages and other sources written partly by Titans of R. The reason for collecting them here, was primarily to have them consolidated in ONE instead of dozens of packages (which themselves might depend on other packages which are not needed at all), and to provide a common and consistent interface as far as function and arguments naming, NA handling, recycling rules etc. are concerned. Google style guides were used as naming rules (in absence of convincing alternatives). The 'camel style' was consequently applied to functions borrowed from contributed R packages as well.
http://cran.r-project.org/web/packages/DescTools/index.html
DescTools contains a bunch of basic statistic functions and convenience wrappers for efficiently describing data, creating specific plots, doing reports using MS Word, Excel or PowerPoint. The package's intention is to offer a toolbox, which facilitates the (notoriously time consuming) first descriptive tasks in data analysis, consisting of calculating descriptive statistics, drawing graphical summaries and reporting the results. Many of the included functions can be found scattered in other packages and other sources written partly by Titans of R. The reason for collecting them here, was primarily to have them consolidated in ONE instead of dozens of packages (which themselves might depend on other packages which are not needed at all), and to provide a common and consistent interface as far as function and arguments naming, NA handling, recycling rules etc. are concerned. Google style guides were used as naming rules (in absence of convincing alternatives). The 'camel style' was consequently applied to functions borrowed from contributed R packages as well.
Freitag, 6. Dezember 2013
Wordcloud
This code yields interesting insights:
library(wordcloud)
txt <- c("hrcds_b","amb_spit_nvl_b","hpp_fg","spez_kons_n","treu_fg",
"qz_n","notfall_n","stat_pv_bl_b","vp_at_p","hosp_n","weibl_p")
n <- c(0.247,0.167,0.084,0.04,
-0.009,-0.012,-0.062,-0.14,-0.143,-0.229,-0.363)
txt <- Sort(data.frame(word=txt, freq=abs(n*1000),
col=PalHelsana()[c("rot", "hellblau")][(n>0)*1+1],
stringsAsFactors=FALSE),2, decr=TRUE)
wordcloud(txt$word, txt$freq, colors=txt$col, ordered.colors=TRUE)
library(wordcloud)
txt <- c("hrcds_b","amb_spit_nvl_b","hpp_fg","spez_kons_n","treu_fg",
"qz_n","notfall_n","stat_pv_bl_b","vp_at_p","hosp_n","weibl_p")
n <- c(0.247,0.167,0.084,0.04,
-0.009,-0.012,-0.062,-0.14,-0.143,-0.229,-0.363)
txt <- Sort(data.frame(word=txt, freq=abs(n*1000),
col=PalHelsana()[c("rot", "hellblau")][(n>0)*1+1],
stringsAsFactors=FALSE),2, decr=TRUE)
wordcloud(txt$word, txt$freq, colors=txt$col, ordered.colors=TRUE)
Montag, 4. November 2013
How to operate in parallel
Doing parallel calculations will spare you much time... There are three points deserving our attention. %dopar% does parallel calculation, the combining function after having the work done can be defined, either c or rbind or whatever and - last but not least - the packages used within the loop must be defined in the specific parameter.
library(doParallel)
cl <- makeCluster(3) # the number of cores to be used
registerDoParallel(cl)
getDoParWorkers() # are they ready?
# remind defining packages if they're used within the loop
res <- foreach(i=1:5, .combine=c, .packages="DescTools") %dopar% {
Primes(i)
}
res
stopCluster(cl) # release your slaves again
Boot already has native support for parallel working:
library(boot)
slopeFun <- function(df, i) {
#df must be a data frame.
#i is the vector of row indices that boot will pass
xResamp <- df[i, ]
slope <- lm(hp ~ cyl + disp, data=xResamp)$coef[2]
}
ptime <- system.time({
b <- boot(mtcars, slopeFun, R=50000, ncpus=6, parallel="snow")
})[3]
ptime
library(doParallel)
cl <- makeCluster(3) # the number of cores to be used
registerDoParallel(cl)
getDoParWorkers() # are they ready?
# remind defining packages if they're used within the loop
res <- foreach(i=1:5, .combine=c, .packages="DescTools") %dopar% {
Primes(i)
}
res
stopCluster(cl) # release your slaves again
Boot already has native support for parallel working:
library(boot)
slopeFun <- function(df, i) {
#df must be a data frame.
#i is the vector of row indices that boot will pass
xResamp <- df[i, ]
slope <- lm(hp ~ cyl + disp, data=xResamp)$coef[2]
}
ptime <- system.time({
b <- boot(mtcars, slopeFun, R=50000, ncpus=6, parallel="snow")
})[3]
ptime
Dienstag, 27. August 2013
Tables in R
We have quite a few table functions in R, all of them solving a specific problem.
library(DescTools)
# Let's define a 3-dim table
# source: http://www.math.wpi.edu/saspdf/stat/chap28.pdf, page 1248
school <- as.table(array(c(35,29, 14,27, 32,10, 53,23), dim=c(2,2,2),
dimnames=list(enrollment=c("yes","no"),
internship=c("yes","no"), gender=c("boys","girls"))))
school
# the percentages
prop.table(school)
# printed as flat table
ftable(school, row.vars=c(3,2))
# aggregate and layout
xtabs(Freq ~ enrollment + gender, data=school)
# and the inverse Operation
tab <- xtabs(Freq ~ enrollment + gender, data=school)
d.frm <- as.data.frame.table(tab)
# factor form with frequencies
data.frame(school)
# get the raw data back in a data.frame
head(Untable(school))
# describe with bells and whistles
Desc(xtabs(Freq ~ internship + enrollment, data=school) )
Assocs(xtabs(Freq ~ internship + enrollment, data=school) )
# how to make a data.frame out of a table
tab <- table(d.pizza$driver, d.pizza$area)
d.frm <- data.frame(unclass(tab))
# or even simpler with base function:
d.frm <- as.data.frame.matrix(tab)
# combine several arrays:
abs <- apply(Titanic, 1:length(dim(Titanic)), formatC, width=3)
rel <- apply(prop.table(Titanic), 1:length(dim(Titanic)), FormatFix, after=3)
ftable(abind(abs, rel))
library(DescTools)
# Let's define a 3-dim table
# source: http://www.math.wpi.edu/saspdf/stat/chap28.pdf, page 1248
school <- as.table(array(c(35,29, 14,27, 32,10, 53,23), dim=c(2,2,2),
dimnames=list(enrollment=c("yes","no"),
internship=c("yes","no"), gender=c("boys","girls"))))
school
# the percentages
prop.table(school)
# printed as flat table
ftable(school, row.vars=c(3,2))
# aggregate and layout
xtabs(Freq ~ enrollment + gender, data=school)
# and the inverse Operation
tab <- xtabs(Freq ~ enrollment + gender, data=school)
d.frm <- as.data.frame.table(tab)
# factor form with frequencies
data.frame(school)
# get the raw data back in a data.frame
head(Untable(school))
# describe with bells and whistles
Desc(xtabs(Freq ~ internship + enrollment, data=school) )
Assocs(xtabs(Freq ~ internship + enrollment, data=school) )
# how to make a data.frame out of a table
tab <- table(d.pizza$driver, d.pizza$area)
d.frm <- data.frame(unclass(tab))
# or even simpler with base function:
d.frm <- as.data.frame.matrix(tab)
# combine several arrays:
abs <- apply(Titanic, 1:length(dim(Titanic)), formatC, width=3)
rel <- apply(prop.table(Titanic), 1:length(dim(Titanic)), FormatFix, after=3)
ftable(abind(abs, rel))
Donnerstag, 13. Juni 2013
Plot intersecting areas
Ever had to plot intersecting areas?
From now on I would use rgeos to do similar things. The following does the trick:
library(rgeos)
library(DescTools)
Canvas(, xpd=TRUE)
p1 <- as(DrawCircle(0.5)[[1]], "gpc.poly")
p2 <- as(DrawCircle(-0.5,0.5)[[1]], "gpc.poly")
p3 <- as(DrawCircle(0,-0.5)[[1]], "gpc.poly")
plot(append.poly(append.poly(p1, p2), p3), axes=FALSE, frame.plot=FALSE, xlab="", ylab="")
plot(intersect(p1, p2), poly.args = list(col = "red"), add = TRUE)
plot(intersect(intersect(p1, p2), p3), poly.args = list(col = "blue"), add = TRUE)
From now on I would use rgeos to do similar things. The following does the trick:
library(rgeos)
library(DescTools)
Canvas(, xpd=TRUE)
p1 <- as(DrawCircle(0.5)[[1]], "gpc.poly")
p2 <- as(DrawCircle(-0.5,0.5)[[1]], "gpc.poly")
p3 <- as(DrawCircle(0,-0.5)[[1]], "gpc.poly")
plot(append.poly(append.poly(p1, p2), p3), axes=FALSE, frame.plot=FALSE, xlab="", ylab="")
plot(intersect(p1, p2), poly.args = list(col = "red"), add = TRUE)
plot(intersect(intersect(p1, p2), p3), poly.args = list(col = "blue"), add = TRUE)
Samstag, 20. April 2013
Second axis
Just a short snippet for a plot with 2 axes for demonstrating the technique.
b <- barplot(1:10, ylim=c(0,20))
par(new=TRUE)
plot(x=b, y=rep(60, 10), xlim=par("usr")[1:2], xaxs="i", yaxs="i"
, frame.plot=FALSE, axes=FALSE, xlab="", ylab="", type="b", pch=15
, ylim=c(0,80))
axis(side=4)
And the result:
b <- barplot(1:10, ylim=c(0,20))
par(new=TRUE)
plot(x=b, y=rep(60, 10), xlim=par("usr")[1:2], xaxs="i", yaxs="i"
, frame.plot=FALSE, axes=FALSE, xlab="", ylab="", type="b", pch=15
, ylim=c(0,80))
axis(side=4)
And the result:
Dienstag, 4. Dezember 2012
Barplot in Tibco Style
Tibco has a nice software which produces good looking plots:
 tibco.colors <- apply( mcol <- matrix(c(
tibco.colors <- apply( mcol <- matrix(c(
0,91,0, 0,157,69, 253,1,97, 60,120,177,
156,205,36, 244,198,7, 254,130,1,
96,138,138, 178,113,60
), ncol=3, byrow=TRUE), 1, function(x) rgb(x[1], x[2], x[3], max=255))
x <- c(12,8,6,4,3,1,1)
par(mgp=c(2.2,0.7,0) ) # decrease distance label to axis
b <- barplot( x, col=rev(tibco.colors), main="Tibco style", border=NA, cex.axis=0.7, las=1, ylim=c(0,13), yaxt="n")
text( b, x+0.6, label=x, cex=0.7)
axis(side=1, at=b, labels=c("EX","VIT","EC","CE","LA","CO","PA"), cex.axis=0.7, las=2, col="grey", las=1, tck=0, xaxs="i")
abline(h=0, col="grey") # just to ensure the axis going through (0,0)
# find the centers of the bars and the gaps
run.mean <- filter( b, filter=c(0.5,0.5))[-length(b)]
gapx <- c(run.mean[1]-diff(b)[1], run.mean, run.mean+diff(b) )
rug(gapx, -0.025, col="grey")
axis(side=2, at=0:12, labels=0:12, cex.axis=0.7, las=2, col="grey", las=1, tck=-0.025)
legend(x="topright", legend=c("Marking","","CONMEDS","DEMOGRAPHICS","ECG","EXPOSURE","LAB_RESULT")
, fill=c(tibco.colors[1],NA,tibco.colors[2:10]), cex=0.7, box.col=NA, border=c("black",NA,rep("black",10)))
 tibco.colors <- apply( mcol <- matrix(c(
tibco.colors <- apply( mcol <- matrix(c(0,91,0, 0,157,69, 253,1,97, 60,120,177,
156,205,36, 244,198,7, 254,130,1,
96,138,138, 178,113,60
), ncol=3, byrow=TRUE), 1, function(x) rgb(x[1], x[2], x[3], max=255))
x <- c(12,8,6,4,3,1,1)
par(mgp=c(2.2,0.7,0) ) # decrease distance label to axis
b <- barplot( x, col=rev(tibco.colors), main="Tibco style", border=NA, cex.axis=0.7, las=1, ylim=c(0,13), yaxt="n")
text( b, x+0.6, label=x, cex=0.7)
axis(side=1, at=b, labels=c("EX","VIT","EC","CE","LA","CO","PA"), cex.axis=0.7, las=2, col="grey", las=1, tck=0, xaxs="i")
# find the centers of the bars and the gaps
run.mean <- filter( b, filter=c(0.5,0.5))[-length(b)]
gapx <- c(run.mean[1]-diff(b)[1], run.mean, run.mean+diff(b) )
rug(gapx, -0.025, col="grey")
axis(side=2, at=0:12, labels=0:12, cex.axis=0.7, las=2, col="grey", las=1, tck=-0.025)
legend(x="topright", legend=c("Marking","","CONMEDS","DEMOGRAPHICS","ECG","EXPOSURE","LAB_RESULT")
, fill=c(tibco.colors[1],NA,tibco.colors[2:10]), cex=0.7, box.col=NA, border=c("black",NA,rep("black",10)))
Samstag, 17. November 2012
dots arguments
How can we parse the dots-arguments in a function?
dots <- function(...){
# was cex in the dots-args? parse dots.arguments
cex <- unlist(match.call(expand.dots=FALSE)$...["cex"])
if(is.null(cex)) print("No cex supplied")
else print(gettextf("cex was %s", cex))
}
dots (cex=0.9, another=4)
dots (another=5)
dots <- function(...){
# was cex in the dots-args? parse dots.arguments
cex <- unlist(match.call(expand.dots=FALSE)$...["cex"])
if(is.null(cex)) print("No cex supplied")
else print(gettextf("cex was %s", cex))
}
dots (cex=0.9, another=4)
dots (another=5)
Donnerstag, 4. Oktober 2012
Get all loaded data.frames
Which data.frames have we loaded?
ls()[ lapply( lapply(ls(), function(x) gettextf("class(%s)", x)), function(x) eval(parse(text=x))) == "data.frame" ]
All available functions in a package:
ls.str(pos = 1, mode = "function")
All installed packages:
search()
(.packages())
(installed.packages())
All available function on search() path:
strsplit( lsf.str(pos = length(search())), " :")
ls()[ lapply( lapply(ls(), function(x) gettextf("class(%s)", x)), function(x) eval(parse(text=x))) == "data.frame" ]
All available functions in a package:
ls.str(pos = 1, mode = "function")
All installed packages:
search()
(.packages())
(installed.packages())
All available function on search() path:
strsplit( lsf.str(pos = length(search())), " :")
Dienstag, 18. September 2012
Conditional Mosaics: Cotabplot
Fine implementation, thanks to Achim Zeileis.
library(vcd)
data("Punishment", package = "vcd")
pun <- xtabs(Freq ~ memory + attitude + age + education, data = Punishment)
cotabplot(~ memory + attitude | age + education, data = pun, panel = cotab_coindep,
n = 5000, type = "mosaic", test = "maxchisq", interpolate = 1:2 )
library(vcd)
data("Punishment", package = "vcd")
pun <- xtabs(Freq ~ memory + attitude + age + education, data = Punishment)
cotabplot(~ memory + attitude | age + education, data = pun, panel = cotab_coindep,
n = 5000, type = "mosaic", test = "maxchisq", interpolate = 1:2 )
Donnerstag, 13. September 2012
Conversions
How can we split a matrix columnwise to a list?
m <- matrix(rnorm(10 * 2), ncol = 2)
The first solution uses a double conversion via a data.frame:
as.list(as.data.frame(m))
The second uses split:
split(x=t(m), f=1:ncol(m))
The third solution works with lapply:
lapply( seq_len(ncol(m)), function(k) m[,k])
m <- matrix(rnorm(10 * 2), ncol = 2)
The first solution uses a double conversion via a data.frame:
as.list(as.data.frame(m))
The second uses split:
split(x=t(m), f=1:ncol(m))
The third solution works with lapply:
lapply( seq_len(ncol(m)), function(k) m[,k])
Dienstag, 4. September 2012
Flexible arguments for R functions
How can we remain flexible concerning arguments of an R function used within a user function. Say we want a histogram to be included in our plot and be able to define all of it's arguments.
I found a good solution in some R-Core-code (don't remember exactly where...).
It goes like that:
myFunction <- function( x, main = "", args.hist = NULL, ... ) {
# define default arguments
args.hist1 <- list(x = x, xlab = "", ylab = "", freq = FALSE,
xaxt = "n", xlim = NULL, ylim = NULL, main = main, las = 1,
col = "white", cex.axis = 1.2)
# override default arguments with user defined ones
if (!is.null(args.hist)) {
args.hist1[names(args.hist)] <- args.hist
}
# call function by means of do.call with the list of arguments
# we can even filter the arguments by name here...
res <- do.call("hist", c(args.hist1[names(args.hist1) %in%
c("x", "breaks", "include.lowest", "right", "nclass")],
plot = FALSE))
}
The function call would then be:
myFunction( x=1:10, args.hist = list(right = TRUE) )
So we have a fine control over which arguments we want to be passed (include.lowest, right) and which ones not (ex. col, cex.axis).
I found a good solution in some R-Core-code (don't remember exactly where...).
It goes like that:
myFunction <- function( x, main = "", args.hist = NULL, ... ) {
# define default arguments
args.hist1 <- list(x = x, xlab = "", ylab = "", freq = FALSE,
xaxt = "n", xlim = NULL, ylim = NULL, main = main, las = 1,
col = "white", cex.axis = 1.2)
# override default arguments with user defined ones
if (!is.null(args.hist)) {
args.hist1[names(args.hist)] <- args.hist
}
# call function by means of do.call with the list of arguments
# we can even filter the arguments by name here...
res <- do.call("hist", c(args.hist1[names(args.hist1) %in%
c("x", "breaks", "include.lowest", "right", "nclass")],
plot = FALSE))
}
The function call would then be:
myFunction( x=1:10, args.hist = list(right = TRUE) )
So we have a fine control over which arguments we want to be passed (include.lowest, right) and which ones not (ex. col, cex.axis).
Mittwoch, 16. Mai 2012
Find all subsets
How can we get all subsets of a vector's elements?
all.subsets <- function(x, min.n=1, max.n=length(x)){
# return a list with all subsets of x
# Caution: This can be heavy for moderate lengths of x11
lst <- lapply( min.n:max.n, function(i) {
m <- combn(x,i) # this is a matrix, split into it's columns
lapply( seq_len(ncol(m)), function(k) m[,k])
} )
# Alternative:
# lst <- lapply(min.n:max.n, function(i) lapply(apply(combn(x,i),2,list),unlist))
# and now flatten the list of lists into one list
lst <- split(unlist(lst), rep(1:length(idx <- rapply(lst, length)), idx))
return(lst)
}
# example:
y <- letters[1:5]
all.subsets(y)
all.subsets <- function(x, min.n=1, max.n=length(x)){
# return a list with all subsets of x
# Caution: This can be heavy for moderate lengths of x11
lst <- lapply( min.n:max.n, function(i) {
m <- combn(x,i) # this is a matrix, split into it's columns
lapply( seq_len(ncol(m)), function(k) m[,k])
} )
# Alternative:
# lst <- lapply(min.n:max.n, function(i) lapply(apply(combn(x,i),2,list),unlist))
# and now flatten the list of lists into one list
lst <- split(unlist(lst), rep(1:length(idx <- rapply(lst, length)), idx))
return(lst)
}
# example:
y <- letters[1:5]
all.subsets(y)
Dienstag, 8. Mai 2012
Run all examples of a package
Get all the base and recommended packages:
installed.packages(priority = c("base","recommended"))
A list of the functions in a package can easily be created by:
ls(pos = "package:MASS")
Run the examples for the first 10 functions of the package MASS:
for( x in paste("example(`", ls(pos="package:MASS")[1:10],"`)", sep="")){
eval(parse(text=x))
}
installed.packages(priority = c("base","recommended"))
A list of the functions in a package can easily be created by:
ls(pos = "package:MASS")
Run the examples for the first 10 functions of the package MASS:
for( x in paste("example(`", ls(pos="package:MASS")[1:10],"`)", sep="")){
eval(parse(text=x))
}
Freitag, 23. März 2012
Running mean, median and others
The running mean (moving average) can be calculated in R by means of the function
x <- c(1,2,4,2,3,4,2,3)
filter(x, rep(1/3,3) )
and the running median by
runmed(x,3)
sequential differences by
diff(x)
for other statistics use the library zoo (and there maybe rollapply).
The time series functions can also be useful for similar problems (e.g. deltat, cycle).
package {caTools} has implemented some fast algorithms for similar purposes:
runmean(x), rumin(x), runmax(x), runquantile(x), runmad(x), runsd(x)
x <- c(1,2,4,2,3,4,2,3)
filter(x, rep(1/3,3) )
and the running median by
runmed(x,3)
sequential differences by
diff(x)
for other statistics use the library zoo (and there maybe rollapply).
The time series functions can also be useful for similar problems (e.g. deltat, cycle).
package {caTools} has implemented some fast algorithms for similar purposes:
runmean(x), rumin(x), runmax(x), runquantile(x), runmad(x), runsd(x)
Donnerstag, 15. März 2012
regexpr examples (just to remember)
Find names ending in "_id" or in "_c" (this would be the same as x like any ('%_id', '%_c') in SQL flavour):
x <- c("vp_id","man_id","min_A_d","min_B_d","count_n",
"type_c","birth_d","gender_c","age_n","hist_y")
x[grep("_(id)|c$", x)]
[1] "vp_id" "man_id" "type_c" "gender_c"
Get rid of everything but the digits:
x <- c("485.2.362.q", "222-445", "889 99 8")
gsub(pattern="[[:punct:]]|[[:alpha:]]|[[:blank:]]", replacement="", x)
Extract uppercase words from the beginning of a string following the idea "delete everything which is not uppercase words":
x <- c("RONALD AYLMER Fisher", "CHARLES Pearson", "John Tukey")
sapply(x, function(x) StrTrim(sub(
pattern=sub(pattern="^[A-ZÄÜÖ -]+\\b\\W+\\b", repl="", x=x)
, repl="", x, fixed=TRUE)))
... and the fine link:
http://www.powerbasic.com/support/help/pbcc/regexpr_statement.htm
x <- c("vp_id","man_id","min_A_d","min_B_d","count_n",
"type_c","birth_d","gender_c","age_n","hist_y")
x[grep("_(id)|c$", x)]
[1] "vp_id" "man_id" "type_c" "gender_c"
Get rid of everything but the digits:
x <- c("485.2.362.q", "222-445", "889 99 8")
gsub(pattern="[[:punct:]]|[[:alpha:]]|[[:blank:]]", replacement="", x)
gsub(pattern="[^0-9]", x=x, replacement="")
gsub(pattern="[^[:digit:]]", x=x, replacement="")
Extract uppercase words from the beginning of a string following the idea "delete everything which is not uppercase words":
x <- c("RONALD AYLMER Fisher", "CHARLES Pearson", "John Tukey")
sapply(x, function(x) StrTrim(sub(
pattern=sub(pattern="^[A-ZÄÜÖ -]+\\b\\W+\\b", repl="", x=x)
, repl="", x, fixed=TRUE)))
... and the fine link:
http://www.powerbasic.com/support/help/pbcc/regexpr_statement.htm
Donnerstag, 19. Januar 2012
Swap variables
... just because of it's elegance:
x <- 2; y <- 5
c(x, y)
y <- (x + y) - (x <- y)
c(x, y)
x <- 2; y <- 5
c(x, y)
y <- (x + y) - (x <- y)
c(x, y)
Dienstag, 22. November 2011
Recode a factor
Recoding a factor seems not to be a frequent activity of Ripley & Co. as it is astonishingly laboriously (because of the "else" part of it...).
The shortest trick I found so far is:
x <- factor(sample(letters, 20))
y <- x
levels(y) <- list(
"good"=c("a","b","c"),
"bad"=c("d","e","f"),
"ugly"=c("h","m"),
"else"=setdiff( levels(y), c("a","b","c","d","e","f","h","m") )
)
data.frame(x,y)
Anyone out there a better idea?
The shortest trick I found so far is:
x <- factor(sample(letters, 20))
y <- x
levels(y) <- list(
"good"=c("a","b","c"),
"bad"=c("d","e","f"),
"ugly"=c("h","m"),
"else"=setdiff( levels(y), c("a","b","c","d","e","f","h","m") )
)
data.frame(x,y)
Anyone out there a better idea?
Mittwoch, 19. Oktober 2011
Montag, 19. September 2011
Mengen-Vergleiche
Interessanten Mengenvergleichs-Funktionen:
(A <- c(sort(sample(1:20, 9)),NA))
[1] 1 2 3 12 16 17 18 19 20 NA
(B <- c(sort(sample(3:23, 7)),NA))
[1] 8 10 15 16 18 21 22 NA
# alle Elemente aus A und B (KEINE Duplikate)
union(A, B)
[1] 1 2 3 12 16 17 18 19 20 NA 8 10 15 21 22
# alle Elemente die in A und in B vorkommen (Schnittmenge)
intersect(A, B)
[1] 16 18 NA
# Elemente in A, aber nicht in B vorkommen
setdiff(A, B)
[1] 1 2 3 12 17 19 20
# Elemente in B, aber nicht in A vorkommen
setdiff(B, A)
[1] 8 10 15 21 22
# Enthält A die gleichen Elemente wie B?
setequal(A, B)
[1] FALSE
(A <- c(sort(sample(1:20, 9)),NA))
[1] 1 2 3 12 16 17 18 19 20 NA
(B <- c(sort(sample(3:23, 7)),NA))
[1] 8 10 15 16 18 21 22 NA
# alle Elemente aus A und B (KEINE Duplikate)
union(A, B)
[1] 1 2 3 12 16 17 18 19 20 NA 8 10 15 21 22
# alle Elemente die in A und in B vorkommen (Schnittmenge)
intersect(A, B)
[1] 16 18 NA
# Elemente in A, aber nicht in B vorkommen
setdiff(A, B)
[1] 1 2 3 12 17 19 20
# Elemente in B, aber nicht in A vorkommen
setdiff(B, A)
[1] 8 10 15 21 22
# Enthält A die gleichen Elemente wie B?
setequal(A, B)
[1] FALSE
Donnerstag, 15. September 2011
Function call with a list of arguments
Use do.call to call a function with a previously built list of arguments:
# Define the arguments
args.legend <- list(
x = "topleft"
, legend = c(1,2)
, fill = c("red","blue")
, xjust = 1, yjust = 1)
plot(1:10, 1:10)
# call function with the arguments' list
do.call("legend", args.legend)
# Define the arguments
args.legend <- list(
x = "topleft"
, legend = c(1,2)
, fill = c("red","blue")
, xjust = 1, yjust = 1)
plot(1:10, 1:10)
# call function with the arguments' list
do.call("legend", args.legend)
Get information about invisible built-in functions
How can we see the code of invisible functions, i.e. mosaicplot?
> mosaicplot
yields only:
function (x, ...)
UseMethod("mosaicplot")
but the functions "methods" and "getAnywhere" do the trick:
> methods(mosaicplot)
[1] mosaicplot.default* mosaicplot.formula*
Non-visible functions are asterisked
> getAnywhere("mosaicplot.formula")
A single object matching ‘mosaicplot.formula’ was found
It was found in the following places
registered S3 method for mosaicplot from namespace graphics
namespace:graphics
with value
function (formula, data = NULL, ..., main = deparse(substitute(data)),
subset, na.action = stats::na.omit)
{
main
m <- match.call(expand.dots = FALSE)
.....
> mosaicplot
yields only:
function (x, ...)
UseMethod("mosaicplot")
but the functions "methods" and "getAnywhere" do the trick:
> methods(mosaicplot)
[1] mosaicplot.default* mosaicplot.formula*
Non-visible functions are asterisked
> getAnywhere("mosaicplot.formula")
A single object matching ‘mosaicplot.formula’ was found
It was found in the following places
registered S3 method for mosaicplot from namespace graphics
namespace:graphics
with value
function (formula, data = NULL, ..., main = deparse(substitute(data)),
subset, na.action = stats::na.omit)
{
main
m <- match.call(expand.dots = FALSE)
.....
Donnerstag, 8. September 2011
Layout-Muster
Split screens:
m <- matrix(0:3, 2, 2)
layout(m, c(1, 3), c(1, 3))
layout.show(3)
m <- matrix(1:4, 2, 2)
layout(m, widths=c(1, 3), heights=c(3, 1))
layout.show(4)
m <- matrix(0:3, 2, 2)
layout(m, c(1, 3), c(1, 3))
layout.show(3)
m <- matrix(1:4, 2, 2)
layout(m, widths=c(1, 3), heights=c(3, 1))
layout.show(4)
Date formats and functions
Date formats:
Remember the functions:
d <- as.Date(c("1937-01-10","1916-03-02","1913-09-19","1927-12-23","1947-07-28"))
quarters(d)
[1] "Q1" "Q1" "Q3" "Q4" "Q3"
# ... or as function
quarter <- function (x) {
# Berechnet das Quartal eines Datums
y <- as.numeric( format( x, "%Y") )
paste(y, "Q", (as.POSIXlt(x)$mon)%/%3 + 1, sep = "")
}
# ... or alternatively with cut
cut(d, breaks="quarters")
[1] 1937-01-01 1916-01-01 1913-07-01 1927-10-01 1947-07-01
137 Levels: 1913-07-01 1913-10-01 1914-01-01 1914-04-01 ... 1947-07-01
months(d)
[1] "Januar" "März" "September" "Dezember" "Juli"
format(d,"%B") # months alternative
[1] "Januar" "März" "September" "Dezember" "Juli"
format(d,"%Y") # years
[1] "1937" "1916" "1913" "1927" "1947"
Remember the functions:
d <- as.Date(c("1937-01-10","1916-03-02","1913-09-19","1927-12-23","1947-07-28"))
quarters(d)
[1] "Q1" "Q1" "Q3" "Q4" "Q3"
# ... or as function
quarter <- function (x) {
# Berechnet das Quartal eines Datums
y <- as.numeric( format( x, "%Y") )
paste(y, "Q", (as.POSIXlt(x)$mon)%/%3 + 1, sep = "")
}
# ... or alternatively with cut
cut(d, breaks="quarters")
[1] 1937-01-01 1916-01-01 1913-07-01 1927-10-01 1947-07-01
137 Levels: 1913-07-01 1913-10-01 1914-01-01 1914-04-01 ... 1947-07-01
months(d)
[1] "Januar" "März" "September" "Dezember" "Juli"
format(d,"%B") # months alternative
[1] "Januar" "März" "September" "Dezember" "Juli"
format(d,"%Y") # years
[1] "1937" "1916" "1913" "1927" "1947"
Montag, 4. Juli 2011
Legend variations
par(mar=c(5.1,4.1,4.1,11.1))
plot( x=1:5, y=1:5, type="n", xlab="x", ylab="y" )
legend( x=2, y=6, legend=c("A","B","C")
, fill=c("red","blue","green")
, density=30, bty="n", horiz=TRUE
, xpd=TRUE )
legend( x=2, y=2, xjust=0.5, yjust=0
, title=" My title:", title.col="grey40", title.adj=0
, legend=c("A","B","C","D","E")
, pch=c(22,22,22,45,45), pt.cex=c(1.2,1.2,1.2,2,2)
, col=c(rep("black",3),"orange","red")
, pt.bg=c("blue","green","yellow")
, bg="grey95", cex=0.8
, box.col="darkgrey", box.lwd=3, box.lty="dotted" )
legend("topright", inset=0.05, cex=0.8, bg="white"
, legend=c("A-1","A-2","B-1", "B-2")
, col=c("lightblue","blue","salmon","red"), pch=15, pt.cex=1.5
, y.intersp=1.5, x.intersp=1.5 , ncol=2 )
windowsFonts("sans2"="Arial Black")
usr <- par(font=4, family="sans2" )
legend( x=5.5, y=3, legend=c("Label A","Label B","Label C")
, fill=c("red","orange","yellow")
, border="brown"
, y.intersp=2, text.width=strwidth("Make larger")
, text.col=c("red","orange","yellow")
, xpd=TRUE )
par(usr)
legend( x="bottomleft", inset=0.02, legend=c("A","B","C","D")
, lty=c("dashed","dotted",NA,"solid"), lwd=2, cex=0.8
, pch=c(NA,NA,21,15)
, col=c("red","blue","black","grey"), bg="white" )
Mittwoch, 9. März 2011
merge
A simple merge example:
a <- data.frame(
"id"=c(1,2,3,6,7,8),
"name"=c("Anna","Berta","Claudia","Dora","Eliane","Frida"))
b <- data.frame(
"id"=c(1,2,3,4,5,9),
"ort"=c("Zürich","Davos","Zermatt","Chamonix","Verbier","Sedrun"))
c.inner <- merge(a,b)
c.left <- merge(a,b, all.x=TRUE)
c.right <- merge(a,b, all.y=TRUE)
c.full <- merge(a,b, all.x=TRUE, all.y=TRUE)
a <- data.frame(
"id"=c(1,2,3,6,7,8),
"name"=c("Anna","Berta","Claudia","Dora","Eliane","Frida"))
b <- data.frame(
"id"=c(1,2,3,4,5,9),
"ort"=c("Zürich","Davos","Zermatt","Chamonix","Verbier","Sedrun"))
c.inner <- merge(a,b)
c.left <- merge(a,b, all.x=TRUE)
c.right <- merge(a,b, all.y=TRUE)
c.full <- merge(a,b, all.x=TRUE, all.y=TRUE)
Freitag, 25. Februar 2011
Split - Apply - Combine
Another effort to make groupwise operations with a split - apply - combine solution (maybe we'll need it someday):
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
, typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
, anz=c(5,4,5,8,3,2,9,1) )
# Split to list
groups <- split(d.frm, list(d.frm$name, d.frm$typ))
# Create result vector
results <- vector("list", length(groups))
# Apply
for(i in seq_along(groups)) {
groups[[i]] <- transform(groups[[i]],
rank = rank(-anz, ties.method = "first"))
results[[i]] <- groups[[i]]
}
# Combine
result <- do.call("rbind", results)
result
name typ anz rank
2 Max blau 4 2
4 Max blau 8 1
8 Moritz blau 1 1
3 Max grün 5 1
5 Max grün 3 2
6 Max rot 5 1
61 Moritz rot 2 2
7 Moritz rot 9 1
We might want to try here the elegant function ave as well:
d.frm$rank_g <- ave( -d.frm$anz, d.frm$name, d.frm$typ,
FUN=function(x) rank(x, ties.method="first") )
... and not to forget the specially R-flavour:
split(x, g) <- lapply(split(x, g), FUN)
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
, typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
, anz=c(5,4,5,8,3,2,9,1) )
# Split to list
groups <- split(d.frm, list(d.frm$name, d.frm$typ))
# Create result vector
results <- vector("list", length(groups))
# Apply
for(i in seq_along(groups)) {
groups[[i]] <- transform(groups[[i]],
rank = rank(-anz, ties.method = "first"))
results[[i]] <- groups[[i]]
}
# Combine
result <- do.call("rbind", results)
result
name typ anz rank
2 Max blau 4 2
4 Max blau 8 1
8 Moritz blau 1 1
3 Max grün 5 1
5 Max grün 3 2
6 Max rot 5 1
61 Moritz rot 2 2
7 Moritz rot 9 1
We might want to try here the elegant function ave as well:
d.frm$rank_g <- ave( -d.frm$anz, d.frm$name, d.frm$typ,
FUN=function(x) rank(x, ties.method="first") )
... and not to forget the specially R-flavour:
split(x, g) <- lapply(split(x, g), FUN)
Dienstag, 21. Dezember 2010
Dotchart nach Cleveland
Alternativ zum Barplot, wenn Mittelwerte oder Mediane gruppenweise dargestellt werden sollen:
dotchart( tapply( ToothGrowth$len, list(ToothGrowth$supp, ToothGrowth$dose), mean )
, main="Guinea Pigs' Tooth Growth", cex=0.8
, xlab="Vitamin C dose", ylab="tooth length")
dotchart( tapply( ToothGrowth$len, list(ToothGrowth$supp, ToothGrowth$dose), mean )
, main="Guinea Pigs' Tooth Growth", cex=0.8
, xlab="Vitamin C dose", ylab="tooth length")
Stripchart
Als Alternative zum Boxplot mit wenigen Datenpunkten eignet sich der Stripchart.
stripchart( len ~ dose, data=ToothGrowth, method="jitter", vertical=T
, subset = supp == "VC", col="blue", main="Guinea Pigs' Tooth Growth"
, xlab="Vitamin C dose", ylab="tooth length")
stripchart( len ~ dose, data=ToothGrowth, method="jitter", vertical=T
, subset = supp == "OJ", col="red", add=T)
stripchart( len ~ dose, data=ToothGrowth, method="jitter", vertical=T
, subset = supp == "VC", col="blue", main="Guinea Pigs' Tooth Growth"
, xlab="Vitamin C dose", ylab="tooth length")
stripchart( len ~ dose, data=ToothGrowth, method="jitter", vertical=T
, subset = supp == "OJ", col="red", add=T)
Donnerstag, 11. November 2010
Grösse des Plot-Fensters bestimmen
Die Grösse des Plot-Fensters kann mit windows() festgelegt werden. Das Grafikfenster wird mit dev.off() wieder geschlossen.
windows(width=10, height=5)
plot( 1:5 )
dev.off()
windows(width=10, height=5)
plot( 1:5 )
dev.off()
Mittwoch, 10. November 2010
Selektieren von Listenelementen
Listenelemente können systematisch mit lapply( list, "[", element ) extrahiert werden.
d.frm <- data.frame( id_name=c("1-Max","2-Maria","3-Steven","4-Jane"))
# split into list
lst <- strsplit( x=as.character(d.frm$id_name), split="-" )
# get first elements out of list
d.frm$id <- as.integer( unlist( lapply( lst, "[", 1) ))
d.frm$name <- unlist( lapply( lst, "[", 2) )
d.frm
id_name name id
1 1-Max Max 1
2 2-Maria Maria 2
3 3-Steven Steven 3
4 4-Jane Jane 4
str(d.frm)
'data.frame': 4 obs. of 3 variables:
$ id_name: Factor w/ 4 levels "1-Max","2-Maria",..: 1 2 3 4
$ name : chr "Max" "Maria" "Steven" "Jane"
$ id : int 1 2 3 4
d.frm <- data.frame( id_name=c("1-Max","2-Maria","3-Steven","4-Jane"))
# split into list
lst <- strsplit( x=as.character(d.frm$id_name), split="-" )
# get first elements out of list
d.frm$id <- as.integer( unlist( lapply( lst, "[", 1) ))
d.frm$name <- unlist( lapply( lst, "[", 2) )
d.frm
id_name name id
1 1-Max Max 1
2 2-Maria Maria 2
3 3-Steven Steven 3
4 4-Jane Jane 4
str(d.frm)
'data.frame': 4 obs. of 3 variables:
$ id_name: Factor w/ 4 levels "1-Max","2-Maria",..: 1 2 3 4
$ name : chr "Max" "Maria" "Steven" "Jane"
$ id : int 1 2 3 4
Donnerstag, 23. September 2010
Reduce margin between plot region and axes with xaxs, yaxs
# get some data
x.i <- seq(0,1,length=5); y.i <- c( 0,0.1,0.2,0.8,1)
par(mfrow=c(1,2))
plot( y=y.i, x=x.i, type="s", panel.before=grid())
symbols( x=0, y=0, circles=0.12, inches=F, add=T, xpd=T, bg=rgb(0,0,1,0.2) )
plot( y=y.i, x=x.i, type="s", xaxs="i", yaxs="i")
grid(); box()
symbols( x=0, y=0, circles=0.12, inches=F, add=T, xpd=T, bg=rgb(0,0,1,0.2) )

x.i <- seq(0,1,length=5); y.i <- c( 0,0.1,0.2,0.8,1)
par(mfrow=c(1,2))
plot( y=y.i, x=x.i, type="s", panel.before=grid())
symbols( x=0, y=0, circles=0.12, inches=F, add=T, xpd=T, bg=rgb(0,0,1,0.2) )
plot( y=y.i, x=x.i, type="s", xaxs="i", yaxs="i")
grid(); box()
symbols( x=0, y=0, circles=0.12, inches=F, add=T, xpd=T, bg=rgb(0,0,1,0.2) )

Dienstag, 21. September 2010
Find most frequent elements
# the vector
x <- sample.int( n=10, size=20, replace=TRUE )
# the 3 most frequent elements
names( head( sort(-table(x)), 3 ) )
# the 3 most frequent elements with their frequencies
head( sort(-table(x)), 3 )
x <- sample.int( n=10, size=20, replace=TRUE )
# the 3 most frequent elements
names( head( sort(-table(x)), 3 ) )
# the 3 most frequent elements with their frequencies
head( sort(-table(x)), 3 )
Freitag, 3. September 2010
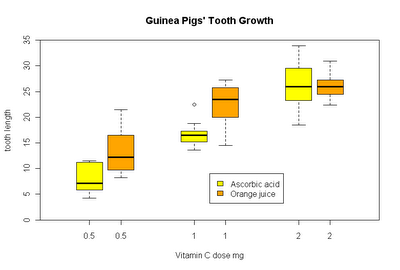
Groupwise boxplot
Groupwise boxplots can easily be created by means of the formula interface.
boxplot(len ~ supp*dose, data = ToothGrowth,
main = "Guinea Pigs' Tooth Growth",
xlab = "Vitamin C dose mg", ylab = "tooth length",
col=c("yellow", "orange")
)
Why an outdated method is described in the boxplot help is however not directly clear. Maybe we are glad to know about the technique anyway someday...
boxplot(len ~ dose, data = ToothGrowth,
boxwex = 0.25, at = 1:3 - 0.15,
subset = supp == "VC", col = "yellow",
main = "Guinea Pigs' Tooth Growth",
xlab = "Vitamin C dose mg",
ylab = "tooth length",
xlim = c(0.5, 3.5), ylim = c(0, 35), yaxs = "i")
boxplot(len ~ dose, data = ToothGrowth, add = TRUE,
boxwex = 0.25, at = 1:3 + 0.15,
subset = supp == "OJ", col = "orange")
legend(2, 9, c("Ascorbic acid", "Orange juice"),
fill = c("yellow", "orange"))
boxplot(len ~ supp*dose, data = ToothGrowth,
main = "Guinea Pigs' Tooth Growth",
xlab = "Vitamin C dose mg", ylab = "tooth length",
col=c("yellow", "orange")
)
Why an outdated method is described in the boxplot help is however not directly clear. Maybe we are glad to know about the technique anyway someday...
boxplot(len ~ dose, data = ToothGrowth,
boxwex = 0.25, at = 1:3 - 0.15,
subset = supp == "VC", col = "yellow",
main = "Guinea Pigs' Tooth Growth",
xlab = "Vitamin C dose mg",
ylab = "tooth length",
xlim = c(0.5, 3.5), ylim = c(0, 35), yaxs = "i")
boxplot(len ~ dose, data = ToothGrowth, add = TRUE,
boxwex = 0.25, at = 1:3 + 0.15,
subset = supp == "OJ", col = "orange")
legend(2, 9, c("Ascorbic acid", "Orange juice"),
fill = c("yellow", "orange"))
SQL-OLAP in R
How to generate SQL-OLAP functions in R:
d.frm <- data.frame( x=rep(1:4,3), g=gl(4,3,labels=letters[1:4]) )
# SQL-OLAP: sum() over (partition by g)
# (more than 1 grouping variables are enumerated like ave(..., g1,g2,g3, FUN=...)):
d.frm$sum_g <- ave( d.frm$x, d.frm$g, FUN=sum )
# same with rank (decreasing):
d.frm$rank_g <- ave( -d.frm$x, d.frm$g, FUN=rank )
d.frm
# get some more data
d.frm <- data.frame(
id=c("p1","p1","p2","p2","p2","p3","p2","p3","p1","p1","p2"),
A=c(0,1,1,1,0,0,0,0,0,0,0),
B=c(1,0,0,0,0,0,0,0,0,0,0),
C=c(0,0,0,0,1,1,1,0,1,1,1)
)
# get rownumber by group, based by original order
d.frm$rownr <- ave( 1:nrow(d.frm), d.frm$id, FUN=order )
# get some groupwise aggregation on more than one column
d.frmby <- data.frame( lapply( d.frm[,-c(1,5)], tapply, d.frm$id, "max", na.rm=TRUE ))
# (see also 'Split - Apply - Combine' post)
d.frm <- data.frame( x=rep(1:4,3), g=gl(4,3,labels=letters[1:4]) )
# SQL-OLAP: sum() over (partition by g)
# (more than 1 grouping variables are enumerated like ave(..., g1,g2,g3, FUN=...)):
d.frm$sum_g <- ave( d.frm$x, d.frm$g, FUN=sum )
# same with rank (decreasing):
d.frm$rank_g <- ave( -d.frm$x, d.frm$g, FUN=rank )
d.frm
# get some more data
d.frm <- data.frame(
id=c("p1","p1","p2","p2","p2","p3","p2","p3","p1","p1","p2"),
A=c(0,1,1,1,0,0,0,0,0,0,0),
B=c(1,0,0,0,0,0,0,0,0,0,0),
C=c(0,0,0,0,1,1,1,0,1,1,1)
)
# get rownumber by group, based by original order
d.frm$rownr <- ave( 1:nrow(d.frm), d.frm$id, FUN=order )
# get some groupwise aggregation on more than one column
d.frmby <- data.frame( lapply( d.frm[,-c(1,5)], tapply, d.frm$id, "max", na.rm=TRUE ))
# (see also 'Split - Apply - Combine' post)
Mittwoch, 18. August 2010
Barplot mit Fehlerbalken
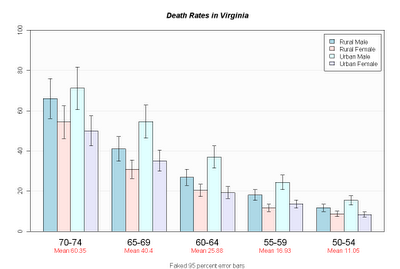
hh <- t(VADeaths)[, 5:1]
ci.l <- hh * 0.85
ci.u <- hh * 1.15
mb <- barplot(hh, beside = TRUE, ylim = c(0, 100)
, col = c("lightblue", "mistyrose","lightcyan", "lavender")
, main = "Death Rates in Virginia", font.main = 4
, sub = "Faked 95 percent error bars", col.sub = "gray20"
, cex.names = 1.5
, legend.text = colnames(VADeaths), args.legend = list( bg="white" )
, panel.before = {
rect( xleft=par()$usr[1], ybottom=par()$usr[3], xright=par()$usr[2], ytop=par()$usr[4]
, col="gray99" )
grid( nx=NA, ny=NULL ) # horiz grid only
box()
}
, xpd=F )
arrows( x0=mb, y0=ci.l, y1 = ci.u, angle=90, code=3, length=0.05 )
mtext( side = 1, at = colMeans(mb), line = 2,
text = paste("Mean", formatC(colMeans(hh))), col = "red" )
Freitag, 6. August 2010
Simple textplot
Putting text on a plot is not that straight on, especially if there's more than one line of text.
data(iris)
# put the summary output into a variable
out <- capture.output(
summary(lm(Sepal.Length ~ Species + Petal.Width, iris)) )
cat( out, sep="\n" )
# create plot
plot.new()
# print text in mono font
text(labels=out, x=0
, y=rev(1:length(out)) * strheight( "S", cex=0.8 ) * 1.3
, adj=c(0,0), family="mono", cex=0.8 )

This is what I thought so far. Of course it is straight on to place bulk text on several lines, stupid...
Just collapse the text with newline as separator:
text(labels=paste(out, collapse="\n"), x=0, y=0
, adj=c(0,0), family="mono", cex=0.8 )
data(iris)
# put the summary output into a variable
out <- capture.output(
summary(lm(Sepal.Length ~ Species + Petal.Width, iris)) )
cat( out, sep="\n" )
# create plot
plot.new()
# print text in mono font
text(labels=out, x=0
, y=rev(1:length(out)) * strheight( "S", cex=0.8 ) * 1.3
, adj=c(0,0), family="mono", cex=0.8 )

This is what I thought so far. Of course it is straight on to place bulk text on several lines, stupid...
Just collapse the text with newline as separator:
text(labels=paste(out, collapse="\n"), x=0, y=0
, adj=c(0,0), family="mono", cex=0.8 )
Dienstag, 18. Mai 2010
Format
Tausender-Trennzeichen und Nachkommastellen
format( 1234, big.mark="'", nsmall=2 )
[1] "1'234.00"
leading zeros
sprintf("%04d", 15)
[1] "0015"
oder mit formatC:
formatC(1:5, width=2, flag="0")
[1] "01" "02" "03" "04" "05"
date
format( Sys.time(), "%Y-%m-%d %H:%M:%S" )
[1] "2010-05-19 10:31:57"
numers as hexnumbers
sprintf("%1$d %1$x %1$X", 0:255)
Codes unter R-Help: strptime
format( 1234, big.mark="'", nsmall=2 )
[1] "1'234.00"
leading zeros
sprintf("%04d", 15)
[1] "0015"
oder mit formatC:
formatC(1:5, width=2, flag="0")
[1] "01" "02" "03" "04" "05"
date
format( Sys.time(), "%Y-%m-%d %H:%M:%S" )
[1] "2010-05-19 10:31:57"
numers as hexnumbers
sprintf("%1$d %1$x %1$X", 0:255)
Codes unter R-Help: strptime
Freitag, 7. Mai 2010
Von R zu Word
Control MS Word from R, do some reporting. The package RDCOMClient is great for that.
The library DescTools (available on CRAN) uses this package and contains some wrapping functions to make things easier.
The library DescTools (available on CRAN) uses this package and contains some wrapping functions to make things easier.
Dienstag, 15. Dezember 2009
List in DataFrame
d.frm <- data.frame(name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
,typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
,anz=c(5,4,5,8,3,2,9,1) )
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm
Zusammenfassung wie summaryBy:
> as.data.frame( do.call("rbind", tapply( d.frm$anz, d.frm$name, summary )))
Min. 1st Qu. Median Mean 3rd Qu. Max.
Max 3 4.0 5 5 5.0 8
Moritz 1 1.5 2 4 5.5 9
,typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
,anz=c(5,4,5,8,3,2,9,1) )
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm
Zusammenfassung wie summaryBy:
> as.data.frame( do.call("rbind", tapply( d.frm$anz, d.frm$name, summary )))
Min. 1st Qu. Median Mean 3rd Qu. Max.
Max 3 4.0 5 5 5.0 8
Moritz 1 1.5 2 4 5.5 9
Dienstag, 17. November 2009
Windows Schriftarten verwenden
Windows Schriftarten müssen definiert werden, bevor sie genutzt werden können:
# Type family examples - creating new mappings
plot(1:10,1:10,type="n")
windowsFonts(
A=windowsFont("Arial Black"),
B=windowsFont("Bookman Old Style"),
C=windowsFont("Comic Sans MS"),
D=windowsFont("Symbol")
)
text(3,3,"Hello World Default")
text(4,4,family="A","Hello World from Arial Black")
text(5,5,family="B","Hello World from Bookman Old Style")
text(6,6,family="C","Hello World from Comic Sans MS")
text(7,7,family="D", "Hello World from Symbol")
# Type family examples - creating new mappings
plot(1:10,1:10,type="n")
windowsFonts(
A=windowsFont("Arial Black"),
B=windowsFont("Bookman Old Style"),
C=windowsFont("Comic Sans MS"),
D=windowsFont("Symbol")
)
text(3,3,"Hello World Default")
text(4,4,family="A","Hello World from Arial Black")
text(5,5,family="B","Hello World from Bookman Old Style")
text(6,6,family="C","Hello World from Comic Sans MS")
text(7,7,family="D", "Hello World from Symbol")
Donnerstag, 5. November 2009
Add a time axis

Adding a time scaled x-axis to a plot can be tedious...
# add a time axis
axis(side = 1, at =as.numeric(seq(as.Date("2000-01-01"), by = "year", length = 10))
, labels = format( seq(as.Date("2000-01-01"), by = "year", length = 10) , "%Y")
, cex.axis = 0.8, las = 1)
# plot the timegrid
# vertical gridlines according to chosen time-values
abline(v = seq(as.Date("2000-01-01"), by = "year", length = 10), col = "grey", lty = "dotted")
# where to plot the horizontal gridlines? get the coords by means of axTicks()
abline(h = axTicks(2), col = "grey", lty = "dotted")
# add a time axis
axis(side = 1, at =as.numeric(seq(as.Date("2000-01-01"), by = "year", length = 10))
, labels = format( seq(as.Date("2000-01-01"), by = "year", length = 10) , "%Y")
, cex.axis = 0.8, las = 1)
# plot the timegrid
# vertical gridlines according to chosen time-values
abline(v = seq(as.Date("2000-01-01"), by = "year", length = 10), col = "grey", lty = "dotted")
# where to plot the horizontal gridlines? get the coords by means of axTicks()
abline(h = axTicks(2), col = "grey", lty = "dotted")
Montag, 5. Oktober 2009
Dummy-Kodierung
Gesucht ist die Dummy-Kodierung eines Faktors:
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
, typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
, anz=c(5,4,5,8,3,2,9,1)
)
d.frm
name typ anz
1 Max rot 5
2 Max blau 4
3 Max grün 5
4 Max blau 8
5 Max grün 3
6 Moritz rot 2
7 Moritz rot 9
8 Moritz blau 1
Gewünschtes liefert
model.matrix(~d.frm$typ)[,-1]
d.frm$typgrün d.frm$typrot
1 0 1
2 0 0
3 1 0
4 0 0
5 1 0
6 0 1
7 0 1
8 0 0
oder alternativ die Funktion class.ind aus der library(nnet):
library(nnet)
class.ind( df$typ )
blau grün rot
[1,] 0 0 1
[2,] 1 0 0
[3,] 0 1 0
[4,] 1 0 0
[5,] 0 1 0
[6,] 0 0 1
[7,] 0 0 1
[8,] 1 0 0
Still another brilliant Ripley solution:
ff <- factor(sample(letters[1:5], 25, replace=TRUE))
diag(nlevels(ff))[ff,]
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
, typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
, anz=c(5,4,5,8,3,2,9,1)
)
d.frm
name typ anz
1 Max rot 5
2 Max blau 4
3 Max grün 5
4 Max blau 8
5 Max grün 3
6 Moritz rot 2
7 Moritz rot 9
8 Moritz blau 1
Gewünschtes liefert
model.matrix(~d.frm$typ)[,-1]
d.frm$typgrün d.frm$typrot
1 0 1
2 0 0
3 1 0
4 0 0
5 1 0
6 0 1
7 0 1
8 0 0
oder alternativ die Funktion class.ind aus der library(nnet):
library(nnet)
class.ind( df$typ )
blau grün rot
[1,] 0 0 1
[2,] 1 0 0
[3,] 0 1 0
[4,] 1 0 0
[5,] 0 1 0
[6,] 0 0 1
[7,] 0 0 1
[8,] 1 0 0
Still another brilliant Ripley solution:
ff <- factor(sample(letters[1:5], 25, replace=TRUE))
diag(nlevels(ff))[ff,]
Freitag, 21. August 2009
Read from/Write to clipboard
Daten lesen aus dem, resp. schreiben in das Clipboard ganz einfach:
d.x <- data.frame( x=c(1,2,3,3), y=c("A","B","A","A") )
write.table(d.x, file="clipboard", sep="\t", row.names=FALSE)
a <- read.table("clipboard", header=TRUE)
d.x <- data.frame( x=c(1,2,3,3), y=c("A","B","A","A") )
write.table(d.x, file="clipboard", sep="\t", row.names=FALSE)
a <- read.table("clipboard", header=TRUE)
Montag, 8. Juni 2009
Modell-Formelnotation
---------------------------------------------------------------------------------------------
Operator Bedeutung
----------------------------------------------------------------------------------------------
+... Hinzunahme einer Variablen
-... Herausnahme einer Variablen (-1 für Achsenabschnitt)
:... Wechselwirkung/Interaktion von Variablen
*... Hinzunahme von Variablen und deren Wechselwirkungen
/... hierarchisch untergeordnet ("nested").
.... y/z bedeutet: z hat nur Wirkung innerhalb der Stufen von y, aber nicht global.
^... alle Interaktionen bis zum angegebenen Grad
.... alle Variablen aus dem Datensatz in das Modell aufnehmen
I(). innerhalb von I() behalten arithmetische Operatoren ihre ursprüngliche Bedeutung
.... ("Inhibit interpretation")
----------------------------------------------------------------------------------------------
Bsp:
y ~ x1 * x2 * x3 - x1:x2:x3 # ohne 3-fach Interaktion
y ~ (x1 + x2 + x3)^2 # alle Variablen inkl. Interaktion bis zum 2. Grad
Operator Bedeutung
----------------------------------------------------------------------------------------------
+... Hinzunahme einer Variablen
-... Herausnahme einer Variablen (-1 für Achsenabschnitt)
:... Wechselwirkung/Interaktion von Variablen
*... Hinzunahme von Variablen und deren Wechselwirkungen
/... hierarchisch untergeordnet ("nested").
.... y/z bedeutet: z hat nur Wirkung innerhalb der Stufen von y, aber nicht global.
^... alle Interaktionen bis zum angegebenen Grad
.... alle Variablen aus dem Datensatz in das Modell aufnehmen
I(). innerhalb von I() behalten arithmetische Operatoren ihre ursprüngliche Bedeutung
.... ("Inhibit interpretation")
----------------------------------------------------------------------------------------------
Bsp:
y ~ x1 * x2 * x3 - x1:x2:x3 # ohne 3-fach Interaktion
y ~ (x1 + x2 + x3)^2 # alle Variablen inkl. Interaktion bis zum 2. Grad
Freitag, 29. Mai 2009
Kombinationen tabellieren
Ausgangslage ist ein data.frame mit einer Zeile pro Element und n Indikatorvariablen. Es stellt sich die Frage nach den Häufigkeiten der Kombinationen der Indikatorvariablen.
d.frm <- data.frame(name=c("Franz", "Maria", "Claudine")
, A=c(1,0,1), B=c(1,1,1), C=c(0,0,1), D=c(0,1,0))
Solution 1 (clumsy):
# Berechne die Summe der mit einer 2er-Potenz multiplizierten Indikatoren
d.frm$sum <- as.matrix(d.frm[,2:5]) %*% (2^(0:3))
# Erzeuge levels für eine Komb-Faktor
d.lvl <- expand.grid( c(0,"A"),c(0,"B"),c(0,"C"),c(0,"D") )
# Bilde eine neue Variable mit der Textbezeichnung der 2^n-Summe
d.frm$sum_x <- factor(d.frm$sum
, levels=((d.lvl!=0)*1) %*% (2^(0:3))
, labels=gsub( "0","", do.call( "paste", c( d.lvl, list(sep="") ))))
d.frm
......name A B C D sum sum_x
1 ...Franz 1 1 0 0 ..3 ...AB
2 ...Maria 0 1 0 1 .10 ...BD
3 Claudine 1 1 1 0 ..7 ..ABC
Solution 2 (smart):
d.frm$sum_xx <- apply( d.frm[,2:5], 1,
function(x) paste(LETTERS[1:4][as.logical(x)], collapse="") )
Wenn einfach nur die Kombinationen gesucht sind, gibt's dafür die Bordmittel:
> combn( letters[1:4], 2 )
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] "a" "a" "a" "b" "b" "c"
[2,] "b" "c" "d" "c" "d" "d"
Oder auch interessant mit den Funktionen outer und lower.tri (pairwise):
m <- outer(x, x, paste, sep="-" )
m[!lower.tri(m, diag=TRUE) ]
[1] "a-b" "a-c" "b-c" "a-d" "b-d" "c-d"
Get all binary combinations
The idea is to get a vector with n 0s and n 1s, chop it into n parts of c(0,1) (this is a list), and use expand.grid:
n <- 4="4">
expand.grid(split(rep(c(0,1), each=n), 1:n))
d.frm <- data.frame(name=c("Franz", "Maria", "Claudine")
, A=c(1,0,1), B=c(1,1,1), C=c(0,0,1), D=c(0,1,0))
Solution 1 (clumsy):
# Berechne die Summe der mit einer 2er-Potenz multiplizierten Indikatoren
d.frm$sum <- as.matrix(d.frm[,2:5]) %*% (2^(0:3))
# Erzeuge levels für eine Komb-Faktor
d.lvl <- expand.grid( c(0,"A"),c(0,"B"),c(0,"C"),c(0,"D") )
# Bilde eine neue Variable mit der Textbezeichnung der 2^n-Summe
d.frm$sum_x <- factor(d.frm$sum
, levels=((d.lvl!=0)*1) %*% (2^(0:3))
, labels=gsub( "0","", do.call( "paste", c( d.lvl, list(sep="") ))))
d.frm
......name A B C D sum sum_x
1 ...Franz 1 1 0 0 ..3 ...AB
2 ...Maria 0 1 0 1 .10 ...BD
3 Claudine 1 1 1 0 ..7 ..ABC
Solution 2 (smart):
d.frm$sum_xx <- apply( d.frm[,2:5], 1,
function(x) paste(LETTERS[1:4][as.logical(x)], collapse="") )
Wenn einfach nur die Kombinationen gesucht sind, gibt's dafür die Bordmittel:
> combn( letters[1:4], 2 )
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] "a" "a" "a" "b" "b" "c"
[2,] "b" "c" "d" "c" "d" "d"
Oder auch interessant mit den Funktionen outer und lower.tri (pairwise):
m <- outer(x, x, paste, sep="-" )
m[!lower.tri(m, diag=TRUE) ]
[1] "a-b" "a-c" "b-c" "a-d" "b-d" "c-d"
Get all binary combinations
The idea is to get a vector with n 0s and n 1s, chop it into n parts of c(0,1) (this is a list), and use expand.grid:
n <- 4="4">
expand.grid(split(rep(c(0,1), each=n), 1:n))
Montag, 25. Mai 2009
Gruppenweise Auswertung
Erstaunlich umständlich ist die gruppenweise Selektion von bestimmten Elementen eines dataframes. Zum Beispiel soll gruppenweise das erste Element nach einer vorgegebenen Sortierung zurückgegeben werden:
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
,typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
,anz=c(5,4,5,8,3,2,9,1) )
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm
....name .typ anz
4 ...Max blau ..8
1 ...Max .rot ..5
3 ...Max grün ..5
2 ...Max blau ..4
5 ...Max grün ..3
7 Moritz .rot ..9
6 Moritz .rot ..2
8 Moritz blau ..1
d.frm[ tapply( rownames(d.frm), d.frm$name, head, n=1), ]
. ..name. typ.anz
4....Max.blau...8
7 Moritz..rot...9
Noch einfacher ist allerdings:
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm[ !duplicated(d.frm$name),]
Wenn der zweite Wert gesucht wäre, ginge dies mit der Index-Funktion "[":
d.frm[ tapply( rownames(d.frm), d.frm$name, "[", 2), ]
d.frm <- data.frame(
name=c("Max","Max","Max","Max","Max","Moritz","Moritz","Moritz")
,typ=c("rot","blau","grün","blau","grün","rot","rot","blau")
,anz=c(5,4,5,8,3,2,9,1) )
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm
....name .typ anz
4 ...Max blau ..8
1 ...Max .rot ..5
3 ...Max grün ..5
2 ...Max blau ..4
5 ...Max grün ..3
7 Moritz .rot ..9
6 Moritz .rot ..2
8 Moritz blau ..1
d.frm[ tapply( rownames(d.frm), d.frm$name, head, n=1), ]
. ..name. typ.anz
4....Max.blau...8
7 Moritz..rot...9
Noch einfacher ist allerdings:
d.frm <- d.frm[ order(d.frm$name, -d.frm$anz),]
d.frm[ !duplicated(d.frm$name),]
Wenn der zweite Wert gesucht wäre, ginge dies mit der Index-Funktion "[":
d.frm[ tapply( rownames(d.frm), d.frm$name, "[", 2), ]
Montag, 6. April 2009
Umorganisation von Daten
Daten, die pivotisiert vorliegen, können mit reshape (siehe auch stack/unstack) wieder in die Faktor-Form gebracht werden.
# Ausgangslage: Breite Form
> d.wide <- data.frame( age=c(22,34,28,31), gender_c=c("m","w","w","m"), ZH=c(12,33,2,5), BE=c(5,6,3,1))
> d.wide
.age gender_c ZH BE
1 22 ....m 12 .5
2 34 ....w 33 .6
3 28 ....w .2 .3
4 31 ....m .5 .1
# ...und die lange Form:
> d.long <- reshape( d.wide, varying=3:4, times=names(d.tmp)[-c(1:2)], v.names="count", direction="long")
> d.long
age gender_c time count id
1.ZH 22 m ZH 12 1
2.ZH 34 w ZH 33 2
3.ZH 28 w ZH 2 3
4.ZH 31 m ZH 5 4
1.BE 22 m BE 5 1
2.BE 34 w BE 6 2
3.BE 28 w BE 3 3
4.BE 31 m BE 1 4
... und wieder zurück:
wide <- reshape( d.long, idvar=c("age","gender_c","id"), timevar="time", direction="wide" )
wide
# Ausgangslage: Breite Form
> d.wide <- data.frame( age=c(22,34,28,31), gender_c=c("m","w","w","m"), ZH=c(12,33,2,5), BE=c(5,6,3,1))
> d.wide
.age gender_c ZH BE
1 22 ....m 12 .5
2 34 ....w 33 .6
3 28 ....w .2 .3
4 31 ....m .5 .1
# ...und die lange Form:
> d.long <- reshape( d.wide, varying=3:4, times=names(d.tmp)[-c(1:2)], v.names="count", direction="long")
> d.long
age gender_c time count id
1.ZH 22 m ZH 12 1
2.ZH 34 w ZH 33 2
3.ZH 28 w ZH 2 3
4.ZH 31 m ZH 5 4
1.BE 22 m BE 5 1
2.BE 34 w BE 6 2
3.BE 28 w BE 3 3
4.BE 31 m BE 1 4
... und wieder zurück:
wide <- reshape( d.long, idvar=c("age","gender_c","id"), timevar="time", direction="wide" )
wide
Sonntag, 15. Februar 2009
Ränder-Problematik

Einstellungen für Ränder in Grafiken für: mar, mai, mgp, omi, oma.
mar=c(5,4,4,2)+0.1 gleich mar=c(u,li,o,re)+0.1,
mar=c(5,4,4,2)+0.1 gleich mar=c(u,li,o,re)+0.1,
mai=c(u,li,o,re) Angabe in inch,
mgp=c(3,1,0) gleich c(Titel, Label, Achse) in mex-Einheiten,
omi=c(0,0,0,0) gleich omi=c(u,li,o,re),
oma=c(0,0,0,0) gleich oma=c(u,li,o,re) in Textzeileneinheiten.
Gauss-Test
Beispiel
Das Gewicht von Briefumschlägen für Luftpost kann als normalverteilt angesehen werden, wobei das mittlere Gewicht weniger als 2 g betragen soll. Die Standardabweichung ist erfahrungsgemäss s=0.02 g. Ein Hersteller entnimmt der Produktion 100 Umschläge und stellt den Mittelwert mw_sp=1.98 g fest.
# Gausstest
n <- 100 # Anzahl Umschläge
MW <- 2 # Mittelwert der Grundgesamtheit
SD <- 0.02 # Standardabweichung der Grundgesamtheit
mw <- 1.98 # Mittelwert der Stichprobe
se <- SD/sqrt(n) # Standardfehler des Mittelwerts
alpha <- 0.05 # Signifikanzniveau
# Quantile der normalverteilung
qnorm(alpha/2, mean=MW, sd=se)
# p-Wert des zweiseitigen Gauss-Testes
2*min(pnorm(mw, mean=MW, sd=se), 1-pnorm(mw, mean=MW, sd=se))
Das Gewicht von Briefumschlägen für Luftpost kann als normalverteilt angesehen werden, wobei das mittlere Gewicht weniger als 2 g betragen soll. Die Standardabweichung ist erfahrungsgemäss s=0.02 g. Ein Hersteller entnimmt der Produktion 100 Umschläge und stellt den Mittelwert mw_sp=1.98 g fest.
# Gausstest
n <- 100 # Anzahl Umschläge
MW <- 2 # Mittelwert der Grundgesamtheit
SD <- 0.02 # Standardabweichung der Grundgesamtheit
mw <- 1.98 # Mittelwert der Stichprobe
se <- SD/sqrt(n) # Standardfehler des Mittelwerts
alpha <- 0.05 # Signifikanzniveau
# Quantile der normalverteilung
qnorm(alpha/2, mean=MW, sd=se)
# p-Wert des zweiseitigen Gauss-Testes
2*min(pnorm(mw, mean=MW, sd=se), 1-pnorm(mw, mean=MW, sd=se))
Abonnieren
Kommentare (Atom)